Input count data
The first step is to input the count data. On the {shiny} app, we can
select to either load the count data from a file (supported formats are
.csv, .dat, and .txt) or to input
the data manually. Once the table is generated and filled, the
“Calculate parameters” button will calculate the total number of cells
(),
total number of aberrations
(),
as well as mean
(),
variance
(),
dispersion index
(),
and
-value.
‘Data input options’ and ‘Fitting options’ boxes in the dose-effect fitting module
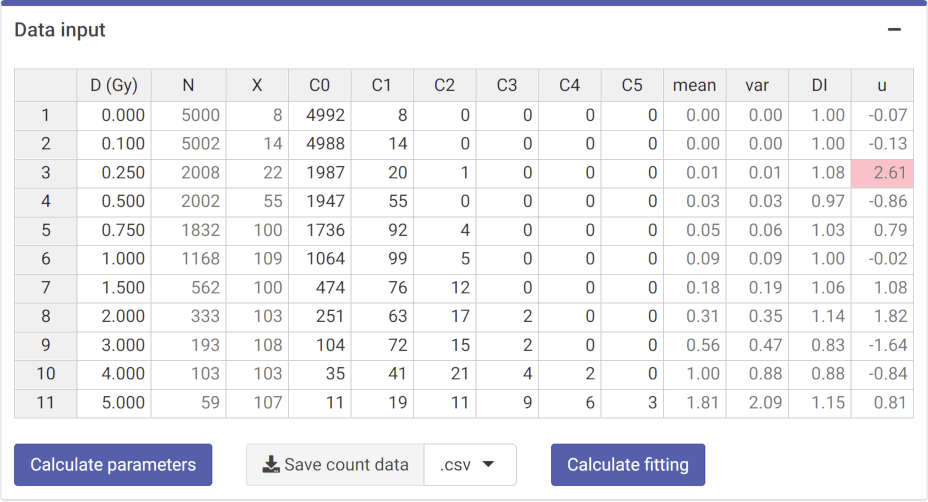
‘Data input’ box in the dose-effect fitting module
This step is accomplished in R by calling the
calculate_aberr_table() function:
count_data <- system.file("extdata", "count-data-barquinero-1995.csv", package = "biodosetools") %>%
utils::read.csv() %>%
calculate_aberr_table(type = "count")
count_data
#> # A tibble: 11 × 13
#> D N X C0 C1 C2 C3 C4 C5 mean var DI
#> <dbl> <int> <dbl> <int> <int> <int> <int> <int> <int> <dbl> <dbl> <dbl>
#> 1 0 5000 8 4992 8 0 0 0 0 0.0016 0.00160 0.999
#> 2 0.1 5002 14 4988 14 0 0 0 0 0.00280 0.00279 0.997
#> 3 0.25 2008 22 1987 20 1 0 0 0 0.0110 0.0118 1.08
#> 4 0.5 2002 55 1947 55 0 0 0 0 0.0275 0.0267 0.973
#> 5 0.75 1832 100 1736 92 4 0 0 0 0.0546 0.0560 1.03
#> 6 1 1168 109 1064 99 5 0 0 0 0.0933 0.0933 0.999
#> 7 1.5 562 100 474 76 12 0 0 0 0.178 0.189 1.06
#> 8 2 333 103 251 63 17 2 0 0 0.309 0.353 1.14
#> 9 3 193 108 104 72 15 2 0 0 0.560 0.466 0.834
#> 10 4 103 103 35 41 21 4 2 0 1 0.882 0.882
#> 11 5 59 107 11 19 11 9 6 3 1.81 2.09 1.15
#> # ℹ 1 more variable: u <dbl>Irradiation conditions
Because irradiation conditions during calibration may influence future dose estimates, and for a better traceability, the user can input the conditions under which the samples used to construct the curve were irradiated. This option is only available in the Shiny app, so that these can be saved into the generated reports.
‘Irradiation conditions’ box in the dose-effect fitting module
Perform fitting
To perform the fitting the user needs to select the appropriate fitting options to click the “Calculate fitting” button on the “Data input” box.
The fitting results and summary statistics are shown in the “Results” tabbed box, and the dose-effect curve is displayed in the “Curve plot” box.
The “Export results” box shows two buttons: (a) “Save fitting data”,
and (b) “Download report”. The “Save fitting data” will generate an
.rds file that contains all information about the count
data, irradiation conditions, and options selected when performing the
fitting. This file can be then loaded in the dose estimation module to
load the dose-effect curve coefficients.
Similarly, the “Download report” will generate a .pdf or
a .docx report containing all inputs and fitting
results.
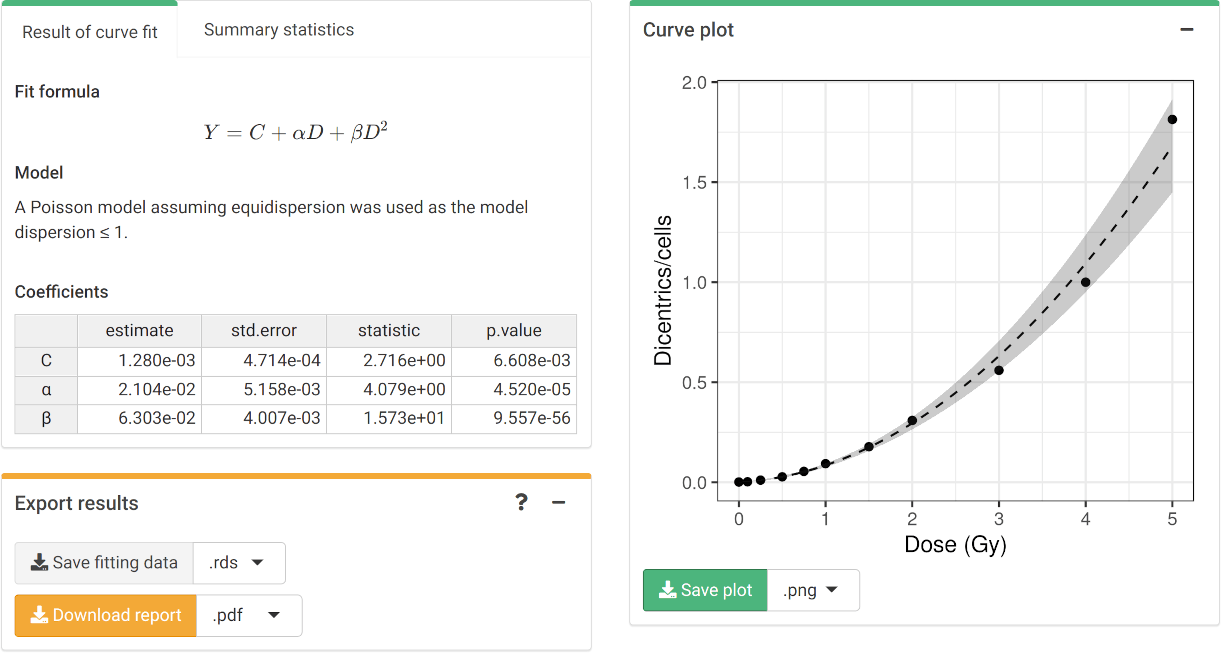
‘Results’ tabbed box, ‘Curve plot’ and ‘Export results’ boxes in the dose-effect fitting module
To perform the fitting in R we call the fit()
function:
fit_results <- fit(
count_data = count_data,
model_formula = "lin-quad",
model_family = "automatic",
fit_link = "identity",
aberr_module = "dicentrics"
)The fit_results object is a list that contains all
necessary information about the count data as well as options selected
when performing the fitting. This is a vital step to ensure traceability
and reproducibility. Below we can see its elements:
names(fit_results)
#> [1] "fit_raw_data" "fit_formula_raw" "fit_formula_tex"
#> [4] "fit_coeffs" "fit_cor_mat" "fit_var_cov_mat"
#> [7] "fit_dispersion" "fit_model_statistics" "fit_algorithm"
#> [10] "fit_model_summary"In particular, we can see how fit_coeffs matches the
results obtained in the UI:
fit_results$fit_coeffs
#> estimate std.error statistic p.value
#> coeff_C 0.001280319 0.0004714055 2.715961 6.608367e-03
#> coeff_alpha 0.021038724 0.0051576170 4.079156 4.519949e-05
#> coeff_beta 0.063032534 0.0040073856 15.729091 9.557291e-56To visualise the dose-effect curve, we call the
plot_fit_dose_curve() function:
plot_fit_dose_curve(
fit_results,
aberr_name = "Dicentrics",
place = "UI"
)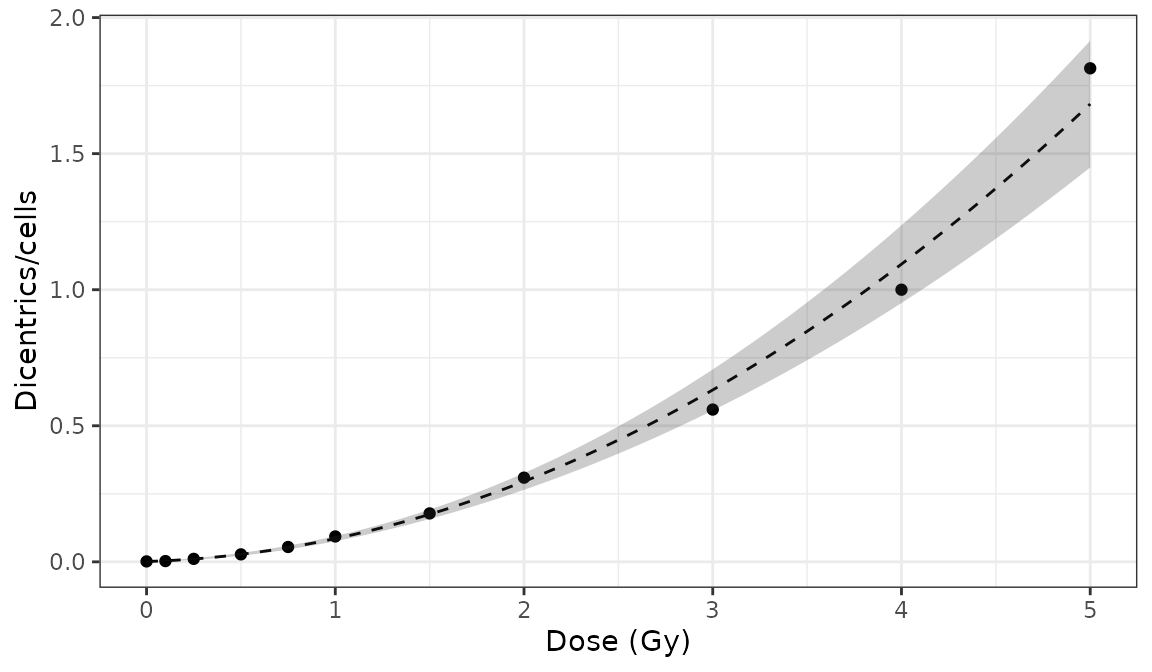
Plot of dose-effect curve generated by {biodosetools}. The grey shading indicates the uncertainties associated with the calibration curve.
