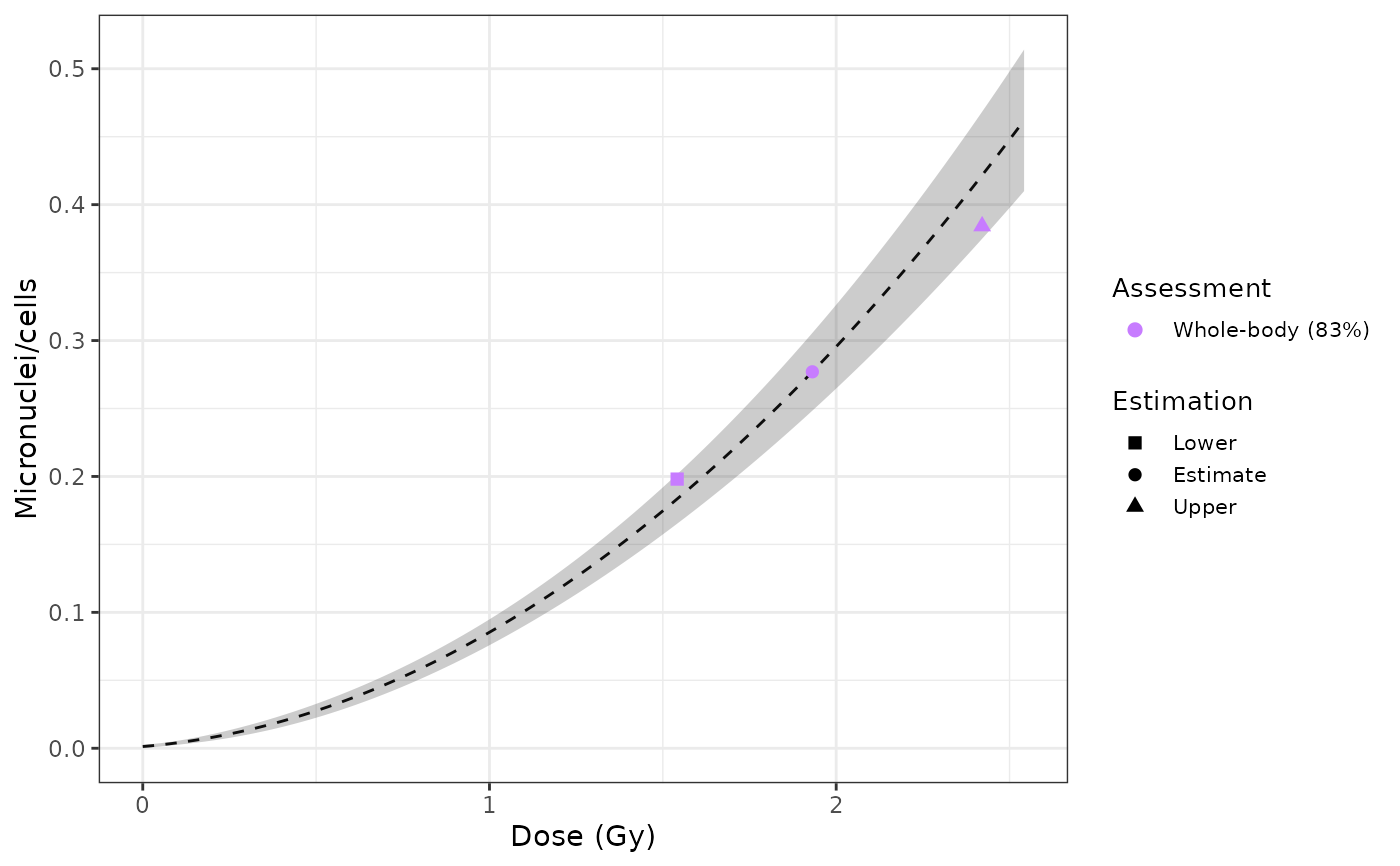
Plot dose estimation curve
Usage
plot_estimated_dose_curve(
est_doses,
fit_coeffs,
fit_var_cov_mat,
protracted_g_value = 1,
conf_int_curve,
aberr_name,
place
)Arguments
- est_doses
List of dose estimations results from
estimate_*()family of functions.- fit_coeffs
Fitting coefficients matrix.
- fit_var_cov_mat
Fitting variance-covariance matrix.
- protracted_g_value
Protracted \(G(x)\) value.
- conf_int_curve
Confidence interval of the curve.
- aberr_name
Name of the aberration to use in the y-axis.
- place
UI or save.
Examples
#The fitting RDS result from the fitting module is needed. Alternatively, manual data
#frames that match the structure of the RDS can be used:
fit_coeffs <- data.frame(
estimate = c(0.001280319, 0.021038724, 0.063032534),
std.error = c(0.0004714055, 0.0051576170, 0.0040073856),
statistic = c(2.715961, 4.079156, 15.729091),
p.value = c(6.608367e-03, 4.519949e-05, 9.557291e-56),
row.names = c("coeff_C", "coeff_alpha", "coeff_beta")
)
fit_var_cov_mat <- data.frame(
coeff_C = c(2.222231e-07, -9.949044e-07, 4.379944e-07),
coeff_alpha = c(-9.949044e-07, 2.660101e-05, -1.510494e-05),
coeff_beta = c(4.379944e-07, -1.510494e-05, 1.605914e-05),
row.names = c("coeff_C", "coeff_alpha", "coeff_beta")
)
results_whole_merkle <- list(
list(
est_doses = data.frame(
lower = 1.541315,
estimate = 1.931213,
upper = 2.420912
),
est_yield = data.frame(
lower = 0.1980553,
estimate = 0.277,
upper = 0.3841655
),
AIC = 7.057229,
conf_int = c(yield_curve = 0.83)
)
)
#FUNCTION PLOT_ESTIMATED_DOSE_CURVE
plot_estimated_dose_curve(
est_doses = list(whole = results_whole_merkle),
fit_coeffs = as.matrix(fit_coeffs),
fit_var_cov_mat = as.matrix(fit_var_cov_mat),
protracted_g_value = 1,
conf_int_curve = 0.95,
aberr_name = "Micronuclei",
place = "UI"
)